목표
- 환자 데이터를 바탕으로 유방암인지 아닌지를 구분해보기
- 딥러닝으로 이진분류 실습을 진행하기
In [1]:
# 라이브러리 불러오기
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer # 사이킷런에 내장되어 있는 유방암 데이터 가져오기
In [2]:
# 데이터 불러오기
breast_data = load_breast_cancer()
print(breast_data)
# 머신러닝 데이터구조: 번치객체(딕셔너리 형태)
{'data': array([[1.799e+01, 1.038e+01, 1.228e+02, ..., 2.654e-01, 4.601e-01,
1.189e-01],
[2.057e+01, 1.777e+01, 1.329e+02, ..., 1.860e-01, 2.750e-01,
8.902e-02],
[1.969e+01, 2.125e+01, 1.300e+02, ..., 2.430e-01, 3.613e-01,
8.758e-02],
...,
[1.660e+01, 2.808e+01, 1.083e+02, ..., 1.418e-01, 2.218e-01,
7.820e-02],
[2.060e+01, 2.933e+01, 1.401e+02, ..., 2.650e-01, 4.087e-01,
1.240e-01],
[7.760e+00, 2.454e+01, 4.792e+01, ..., 0.000e+00, 2.871e-01,
7.039e-02]]), 'target': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0,
1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0,
1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1,
.................................................................
1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1,
1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1]), 'frame': None, 'target_names': array(['malignant', 'benign'], ......................................,
'mean smoothness', 'mean compactness', 'mean concavity',
'mean concave points', 'mean symmetry', 'mean fractal dimension',
'radius error', 'texture error', 'perimeter error', 'area error',
................................................................
'worst compactness', 'worst concavity', 'worst concave points',
'worst symmetry', 'worst fractal dimension'], dtype='</u9'),>In [3]:
breast_data.keys()
Out[3]:
dict_keys(['data', 'target', 'frame', 'target_names', 'DESCR', 'feature_names', 'filename', 'data_module'])In [4]:
# data:문제데이터, 입력특성
# target: 정답데이터
# target_names: 정답데이터의 이름
In [5]:
# 정답데이터 확인
breast_data.target
Out[5]:
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0,
1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0,
1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1,
1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 0,
0, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1,
1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1,
1, 0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0,
0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0,
1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1,
1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0,
0, 1, 1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0,
0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1,
1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 1,
1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0,
1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1,
1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1,
1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1])In [6]:
# 정답데이터 확인
breast_data.target_names
# 0 == 'malignant'(악성), 1 == 'begin'(양성)
# 클래스의 개수: 2개 -> 이진분류
Out[6]:
array(['malignant', 'benign'], dtype='<U9')In [7]:
breast_data.target_names
Out[7]:
array(['malignant', 'benign'], dtype='<U9')In [8]:
# 문제, 정답 분리
# 문제 X, 정답 y
X = breast_data['data'] y = breast_data['target']
In [9]:
# train, test 분리
from sklearn.model_selection import train_test_split
# train_test_split(test_size = 0.2)
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size = 0.2, random_state = 919)
In [10]:
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
(455, 30) (455,)
(114, 30) (114,)
모델링 과정
- 신경망 구조 설계 (뼈대, 입력층, 중간층, 출력층)
- 신경망 모델 학습/평가 방법 설정 (회귀, 분류)
- 모델 학습
- 모델 예측 및 평가
In [11]:
# 모델 생성을 위한 도구 불러오기
from tensorflow.keras.models import Sequential # 뼈대
from tensorflow.keras.layers import InputLayer, Dense
In [12]:
# 1. 모델 구조 설계
# 뼈대
model = Sequential()
# 입력층
model.add(InputLayer(input_shape = (30,))) # 입력층 (입력특성의 개수)
# 중간층(은닉층)
model.add(Dense(units = 16, activation = "sigmoid"))
model.add(Dense(units = 8, activation = "sigmoid"))
# 출력층 출력받고싶은 형태 지정( 이진분류, 0-1사이 확률값 하나)
model.add(Dense(units = 1, activation = "sigmoid"))
In [13]:
# 2. 모델 학습 및 평가 방법 설정
model.compile(loss = 'binary_crossentropy', # 오차 : 이진분류 -> binary_crossentropy
optimizer = 'SGD', # 최적화 알고리즘 (확률적 경사하강법)
metrics = ['accuracy']) # 평가방법(분류:정확도)
In [14]:
h1 = model.fit(X_train, y_train, validation_split = 0.2, epochs = 20)
Epoch 1/20
12/12 [==============================] - 1s 41ms/step - loss: 0.6615 - accuracy: 0.6456 - val_loss: 0.6753 - val_accuracy: 0.5934
Epoch 2/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6585 - accuracy: 0.6456 - val_loss: 0.6716 - val_accuracy: 0.5934
Epoch 3/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6538 - accuracy: 0.6456 - val_loss: 0.6701 - val_accuracy: 0.5934
Epoch 4/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6515 - accuracy: 0.6456 - val_loss: 0.6695 - val_accuracy: 0.5934
Epoch 5/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6500 - accuracy: 0.6456 - val_loss: 0.6687 - val_accuracy: 0.5934
Epoch 6/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6479 - accuracy: 0.6456 - val_loss: 0.6680 - val_accuracy: 0.5934
Epoch 7/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6462 - accuracy: 0.6456 - val_loss: 0.6678 - val_accuracy: 0.5934
Epoch 8/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6448 - accuracy: 0.6456 - val_loss: 0.6662 - val_accuracy: 0.5934
Epoch 9/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6434 - accuracy: 0.6456 - val_loss: 0.6658 - val_accuracy: 0.5934
Epoch 10/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6422 - accuracy: 0.6456 - val_loss: 0.6664 - val_accuracy: 0.5934
Epoch 11/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6413 - accuracy: 0.6456 - val_loss: 0.6648 - val_accuracy: 0.5934
Epoch 12/20
12/12 [==============================] - 0s 5ms/step - loss: 0.6402 - accuracy: 0.6456 - val_loss: 0.6644 - val_accuracy: 0.5934
Epoch 13/20
12/12 [==============================] - 0s 5ms/step - loss: 0.6396 - accuracy: 0.6456 - val_loss: 0.6645 - val_accuracy: 0.5934
Epoch 14/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6387 - accuracy: 0.6456 - val_loss: 0.6634 - val_accuracy: 0.5934
Epoch 15/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6382 - accuracy: 0.6456 - val_loss: 0.6637 - val_accuracy: 0.5934
Epoch 16/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6372 - accuracy: 0.6456 - val_loss: 0.6621 - val_accuracy: 0.5934
Epoch 17/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6365 - accuracy: 0.6456 - val_loss: 0.6625 - val_accuracy: 0.5934
Epoch 18/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6358 - accuracy: 0.6456 - val_loss: 0.6617 - val_accuracy: 0.5934
Epoch 19/20
12/12 [==============================] - 0s 4ms/step - loss: 0.6352 - accuracy: 0.6456 - val_loss: 0.6615 - val_accuracy: 0.5934
Epoch 20/20
12/12 [==============================] - 0s 6ms/step - loss: 0.6343 - accuracy: 0.6456 - val_loss: 0.6605 - val_accuracy: 0.5934
In [15]:
# 결과 시각화
plt.figure(figsize = (15, 5))
plt.plot(h1.history['loss'], label = 'train_loss')
plt.plot(h1.history['val_loss'], label = 'val_loss')
plt.legend() plt.show()
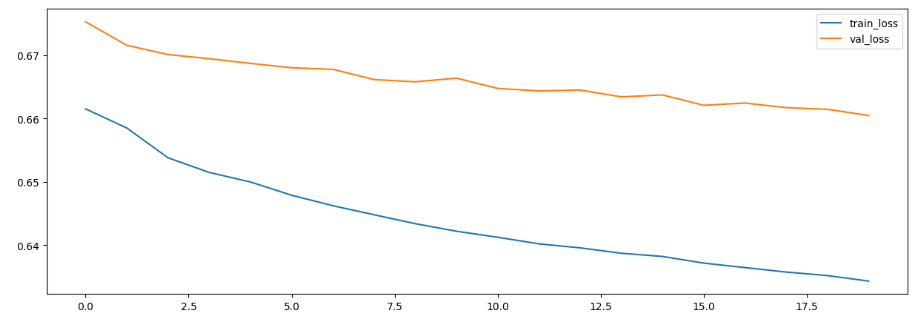
In [16]:
# 모델에 대한 전체적인 내부구조 확인 model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 16) 496
dense_1 (Dense) (None, 8) 136
dense_2 (Dense) (None, 1) 9
=================================================================
Total params: 641 (2.50 KB)
Trainable params: 641 (2.50 KB)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
'Study > DeepLearning' 카테고리의 다른 글
| [DeepLearning] 다중분류 / 손글씨 데이터 분류 실습 (0) | 2023.09.27 |
|---|---|
| [DeepLearning] 딥러닝 시작하기 (0) | 2023.09.25 |

